Import function for WinSpec SPE files (file version up to 3.0). The calibration data (polynomial and calibration data pairs) for x-axis are automatically read and applied to the spectra. Note that the y-calibration data structure is not extracted from the file since it is not saved there by WinSpec and is always empty.
read_spe(
file,
wl_units = NULL,
acc2avg = FALSE,
cts_sec = FALSE,
keys_hdr2data = c("exposure_sec", "LaserWavelen", "accumulCount", "numFrames",
"darkSubtracted"),
xaxis = "DEPRECATED"
)Arguments
- file
(character): Name of the SPE file to read data from.
- wl_units
(character|
NULL): Units of x-axis, e.g.,"px","nm","energy","raman",....NULLmeans default units defined in a file.Function
read_spe()automatically checks if the x-calibration data are available and uses them (if possible) to reconstruct the wavelength axis in the selected units.- acc2avg
(logical): Whether to divide the actual data set by the number of accumulations, thus transforming accumulated spectra to averaged spectra. WinSpec does not do this automatically, so the spectral intensity is always proportional to the number of accumulations. The flag
@data$averagedis automatically set toTRUE.- cts_sec
(logical): Whether to divide the actual data set by the exposure time, thus going to count per second unit.
- keys_hdr2data
(character): Which metadata from the file header should be saved to the
@dataslot of a newly createdhyperSpecobject.- xaxis
(DEPRECATED): use
wl_unitsinstead.
Value
hyperSpec object.
Examples
suppressPackageStartupMessages({
library(hySpc.read.spe)
})
# Example 1
f_blut1 <- system.file("extdata", "blut1.SPE", package = "hySpc.read.spe")
spc_blut1 <- read_spe(f_blut1)
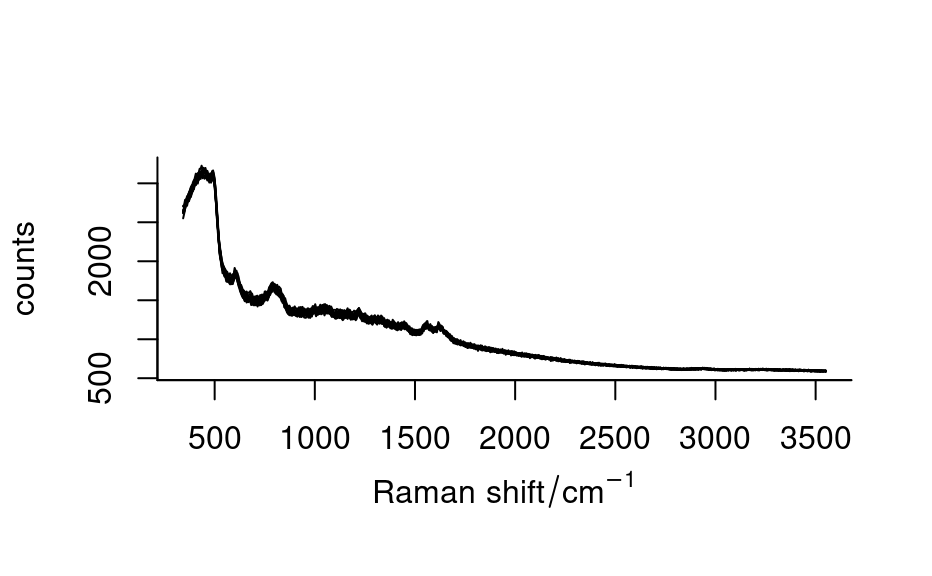
spc_blut1
#> hyperSpec object
#> 10 spectra
#> 9 data columns
#> 1023 data points / spectrum
#> wavelength: Raman ~ shift/cm^-1 [numeric] 342.6594 347.5543 ... 3550.062
#> data: (10 rows x 9 columns)
#> 1. px.y: [integer] 1 1 ... 1
#> 2. frame: [integer] 1 2 ... 10
#> 3. exposure_sec: [numeric] 3 3 ... 3
#> 4. LaserWavelen: [numeric] 784.068 784.068 ... 784.068
#> 5. accumulCount: [integer] 1 1 ... 1
#> 6. numFrames: [integer] 10 10 ... 10
#> 7. darkSubtracted: [integer] 0 0 ... 0
#> 8. spc: counts [matrix, array1023] 2711 2655 ... 597
#> 9. filename: filename [character] /tmp/Rtmp4lLVXr/temp_libpath14eb1e99c560/hySpc.read.spe/extdata/blut1.SPE /tmp/Rtmp4lLVXr/temp_libpath14eb1e99c560/hySpc.read.spe/extdata/blut1.SPE ... /tmp/Rtmp4lLVXr/temp_libpath14eb1e99c560/hySpc.read.spe/extdata/blut1.SPE
plot(spc_blut1)
 # Example 2
f_spe3 <- system.file(
"extdata", "spe_format_3.0.SPE",
package = "hySpc.read.spe"
)
spc_spe3 <- read_spe(f_spe3)
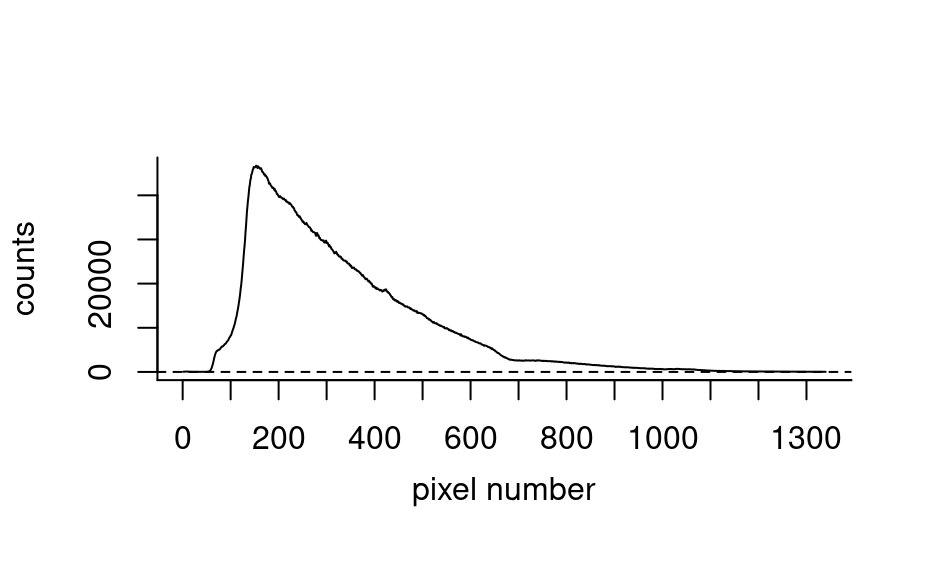
spc_spe3
#> hyperSpec object
#> 1 spectra
#> 10 data columns
#> 1340 data points / spectrum
#> wavelength: pixel number [integer] 1 2 ... 1340
#> data: (1 rows x 10 columns)
#> 1. px.y: [integer] 1
#> 2. frame: [integer] 1
#> 3. exposure_sec: [numeric] 0
#> 4. LaserWavelen: [numeric] 0
#> 5. accumulCount: [integer] 0
#> 6. numFrames: [integer] 1
#> 7. darkSubtracted: [integer] 0
#> 8. spc: counts [matrix, array1340] 38 44 ... 28
#> 9. xml: [list] columns/entries SpeFormat
#> 10. filename: filename [character] /tmp/Rtmp4lLVXr/temp_libpath14eb1e99c560/hySpc.read.spe/extdata/spe_format_3.0.SPE
plot(spc_spe3)
# Example 2
f_spe3 <- system.file(
"extdata", "spe_format_3.0.SPE",
package = "hySpc.read.spe"
)
spc_spe3 <- read_spe(f_spe3)
spc_spe3
#> hyperSpec object
#> 1 spectra
#> 10 data columns
#> 1340 data points / spectrum
#> wavelength: pixel number [integer] 1 2 ... 1340
#> data: (1 rows x 10 columns)
#> 1. px.y: [integer] 1
#> 2. frame: [integer] 1
#> 3. exposure_sec: [numeric] 0
#> 4. LaserWavelen: [numeric] 0
#> 5. accumulCount: [integer] 0
#> 6. numFrames: [integer] 1
#> 7. darkSubtracted: [integer] 0
#> 8. spc: counts [matrix, array1340] 38 44 ... 28
#> 9. xml: [list] columns/entries SpeFormat
#> 10. filename: filename [character] /tmp/Rtmp4lLVXr/temp_libpath14eb1e99c560/hySpc.read.spe/extdata/spe_format_3.0.SPE
plot(spc_spe3)