These functions fit polynomial baselines.
spc_fit_poly(
fit.to,
apply.to = NULL,
poly.order = 1,
offset.wl = !(is.null(apply.to))
)
spc_fit_poly_below(
fit.to,
apply.to = fit.to,
poly.order = 1,
npts.min = max(round(nwl(fit.to) * 0.05), 3 * (poly.order + 1)),
noise = 0,
offset.wl = FALSE,
max.iter = nwl(fit.to),
stop.on.increase = FALSE,
debuglevel = hy_get_option("debuglevel")
)Arguments
- fit.to
hyperSpecobject on which the baselines are fitted- apply.to
hyperSpecobject on which the baselines are evaluted IfNULL, ahyperSpecobject containing the polynomial coefficients rather than evaluted baselines is returned.- poly.order
order of the polynomial to be used
- offset.wl
should the wavelength range be mapped to -> [0, delta wl]? This enhances numerical stability.
- npts.min
minimal number of points used for fitting the polynomial
- noise
noise level to be considered during the fit. It may be given as one value for all the spectra, or for each spectrum separately.
- max.iter
stop at the latest after so many iterations.
- stop.on.increase
additional stopping rule: stop if the number of support points would increase, regardless whether
npts.minwas reached or not.- debuglevel
additional output:
1showsnpts.min,2plots support points for the final baseline of 1st spectrum,3plots support points for 1st spectrum,4plots support points for all spectra.
Value
hyperSpec object containing the baselines in the spectra
matrix, either as polynomial coefficients or as polynomials evaluted on
the spectral range of apply.to
Details
Both functions fit polynomials to be used as baselines. If apply.to
is NULL, a hyperSpec object with the polynomial coefficients
is returned, otherwise the polynomials are evaluated on the spectral range
of apply.to.
spc_fit_poly() calculates the least squares fit of order
poly.order to the complete spectra given in fit.to.
Thus fit.to needs to be cut appropriately.
spc_fit_poly_below() tries to fit the baseline on appropriate spectral
ranges of the spectra in fit.to.
For details, see the vignette("baseline").
See also
vignette("baseline", package = "hyperSpec")
see options() for more on debuglevel
Examples
if (FALSE) {
vignette("baseline", package = "hyperSpec")
}
spc <- faux_cell[1:10]
baselines <- spc_fit_poly(spc[, , c(625 ~ 640, 1785 ~ 1800)], spc)
plot(spc - baselines)
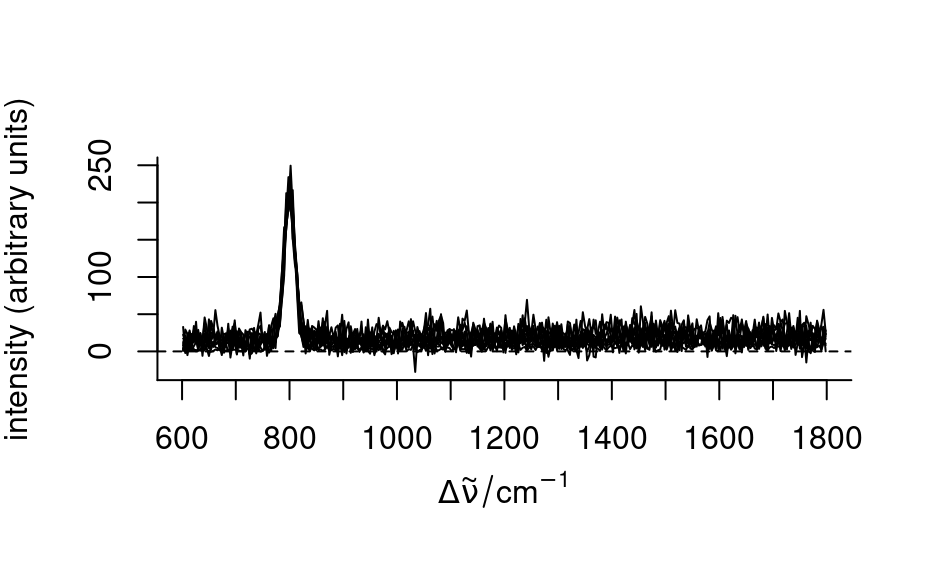
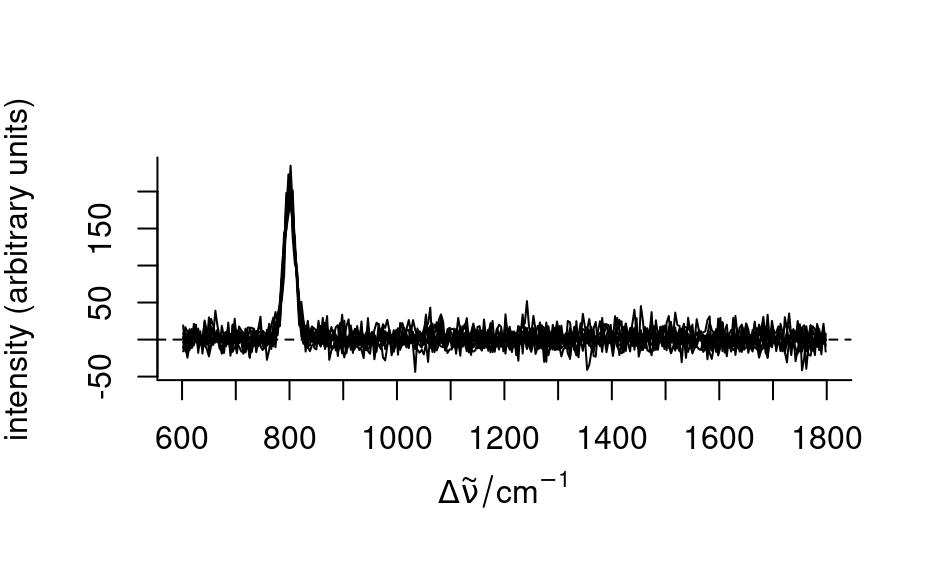 baselines <- spc_fit_poly_below(spc)
plot(spc - baselines)
baselines <- spc_fit_poly_below(spc)
plot(spc - baselines)
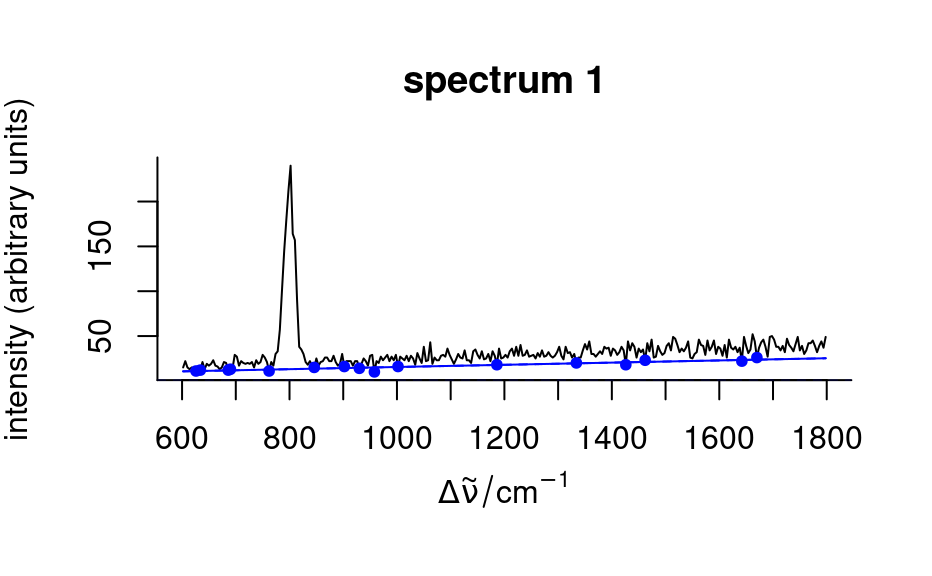
 spc_fit_poly_below(faux_cell[1:3], debuglevel = 1)
#> Fitting with npts.min = 15
#> spectrum 1: 16 support points, noise = 0.0, 5 iterations
#> spectrum 2: 37 support points, noise = 0.0, 4 iterations
#> spectrum 3: 30 support points, noise = 0.0, 4 iterations
#> hyperSpec object
#> 3 spectra
#> 4 data columns
#> 300 data points / spectrum
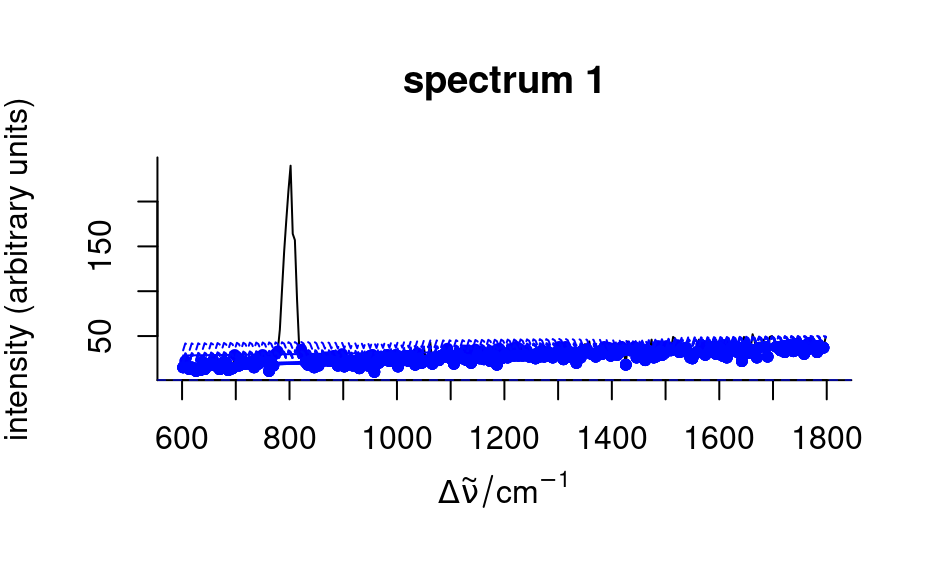
spc_fit_poly_below(faux_cell[1:3], debuglevel = 2)
#> Fitting with npts.min = 15
#> start: 300 support points
#> spectrum 1: 16 support points, noise = 0.0, 5 iterations
#> spectrum 2: 37 support points, noise = 0.0, 4 iterations
#> spectrum 3: 30 support points, noise = 0.0, 4 iterations
spc_fit_poly_below(faux_cell[1:3], debuglevel = 1)
#> Fitting with npts.min = 15
#> spectrum 1: 16 support points, noise = 0.0, 5 iterations
#> spectrum 2: 37 support points, noise = 0.0, 4 iterations
#> spectrum 3: 30 support points, noise = 0.0, 4 iterations
#> hyperSpec object
#> 3 spectra
#> 4 data columns
#> 300 data points / spectrum
spc_fit_poly_below(faux_cell[1:3], debuglevel = 2)
#> Fitting with npts.min = 15
#> start: 300 support points
#> spectrum 1: 16 support points, noise = 0.0, 5 iterations
#> spectrum 2: 37 support points, noise = 0.0, 4 iterations
#> spectrum 3: 30 support points, noise = 0.0, 4 iterations
 #> hyperSpec object
#> 3 spectra
#> 4 data columns
#> 300 data points / spectrum
spc_fit_poly_below(faux_cell[1:3],
debuglevel = 3,
noise = sqrt(rowMeans(faux_cell[[1:3]]))
)
#> Fitting with npts.min = 15
#> start: 300 support points
#> Iteration 1: 260 support points
#> Iteration 2: 229 support points
#> Iteration 3: 216 support points
#> Iteration 4: 213 support points
#> Iteration 5: 212 support points
#> Iteration 6: 212 support points
#> spectrum 1: 212 support points, noise = 5.7, 6 iterations
#> spectrum 2: 201 support points, noise = 13.4, 10 iterations
#> spectrum 3: 181 support points, noise = 11.8, 9 iterations
#> hyperSpec object
#> 3 spectra
#> 4 data columns
#> 300 data points / spectrum
spc_fit_poly_below(faux_cell[1:3],
debuglevel = 3,
noise = sqrt(rowMeans(faux_cell[[1:3]]))
)
#> Fitting with npts.min = 15
#> start: 300 support points
#> Iteration 1: 260 support points
#> Iteration 2: 229 support points
#> Iteration 3: 216 support points
#> Iteration 4: 213 support points
#> Iteration 5: 212 support points
#> Iteration 6: 212 support points
#> spectrum 1: 212 support points, noise = 5.7, 6 iterations
#> spectrum 2: 201 support points, noise = 13.4, 10 iterations
#> spectrum 3: 181 support points, noise = 11.8, 9 iterations
 #> hyperSpec object
#> 3 spectra
#> 4 data columns
#> 300 data points / spectrum
#> hyperSpec object
#> 3 spectra
#> 4 data columns
#> 300 data points / spectrum