Scales the spectra matrix. scale(x, scale = FALSE) centers the data.
# S4 method for hyperSpec
scale(x, center = TRUE, scale = TRUE)Arguments
- x
the
hyperSpecobject- center
if
TRUE, the data is centered tocolMeans(x),FALSEsuppresses centering. Alternatively, an object that can be converted to numeric of lengthnwl(x)bybase::as.matrix()(e.g. hyperSpec object containing 1 spectrum) can specify the center spectrum.- scale
if
TRUE, the data is scaled to have unit variance at each wavelength,FALSEsuppresses scaling. Alternatively, an object that can be converted to numeric of lengthnwl(x)bybase::as.matrix()(e.g. hyperSpec object containing 1 spectrum) can specify the center spectrum.
Value
the centered & scaled hyperSpec object
Details
Package scale provides a fast alternative for base::scale()
See also
package scale.
Examples
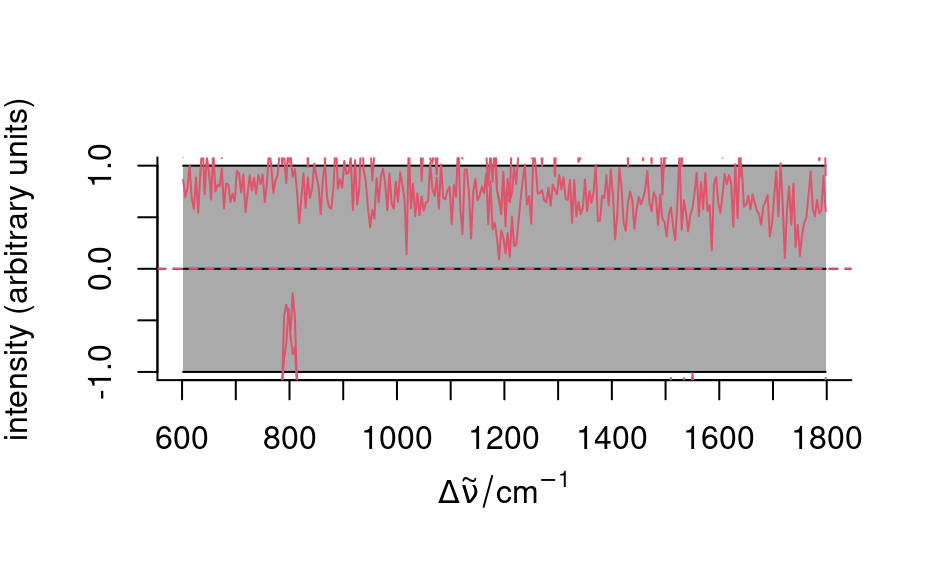
## mean center & variance scale
tmp <- scale(faux_cell)
plot(tmp, "spcmeansd")
plot(sample(tmp, 5), add = TRUE, col = 2)
 ## mean center only
tmp <- scale(faux_cell, scale = FALSE)
plot(tmp, "spcmeansd")
plot(sample(tmp, 5), add = TRUE, col = 2)
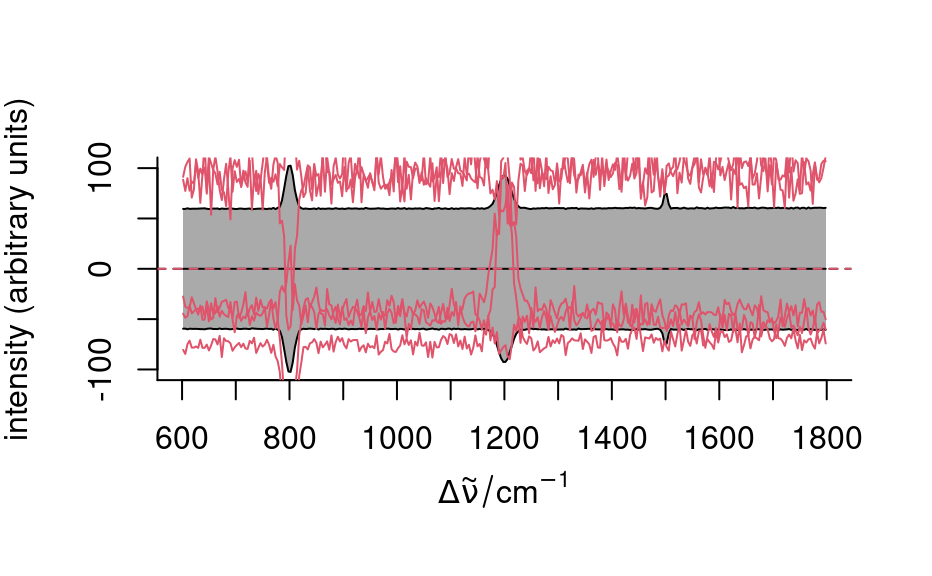
## mean center only
tmp <- scale(faux_cell, scale = FALSE)
plot(tmp, "spcmeansd")
plot(sample(tmp, 5), add = TRUE, col = 2)
 ## custom center
tmp <- sweep(faux_cell, 1, mean, `/`)
plot(tmp, "spcmeansd")
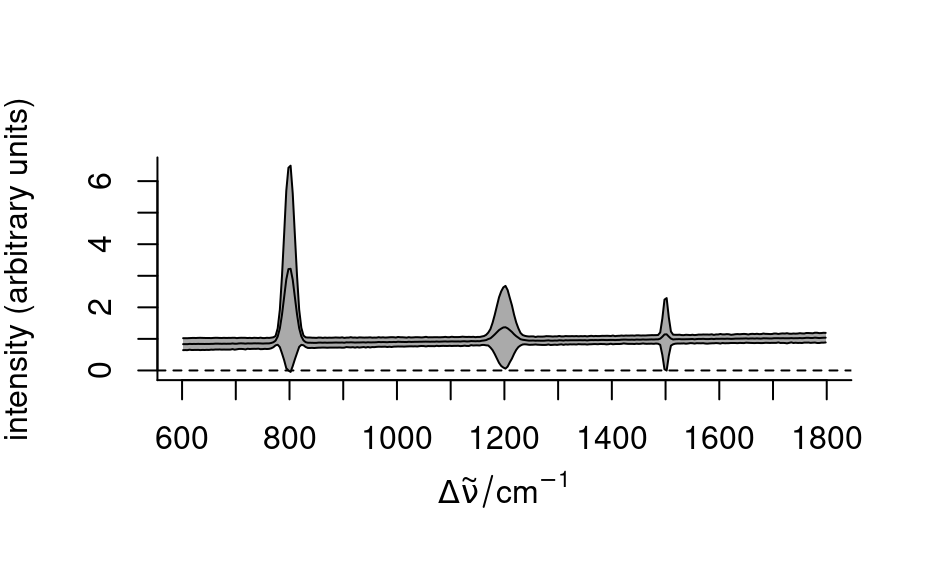
## custom center
tmp <- sweep(faux_cell, 1, mean, `/`)
plot(tmp, "spcmeansd")
 tmp <- scale(tmp, center = quantile(tmp, .05), scale = FALSE)
tmp <- scale(tmp, center = quantile(tmp, .05), scale = FALSE)